♦ Matthias Mann团队深度视觉蛋白质组学培训课程 | AOHUPO&AOAPO&CNHUPO&π-HuB 2025大会
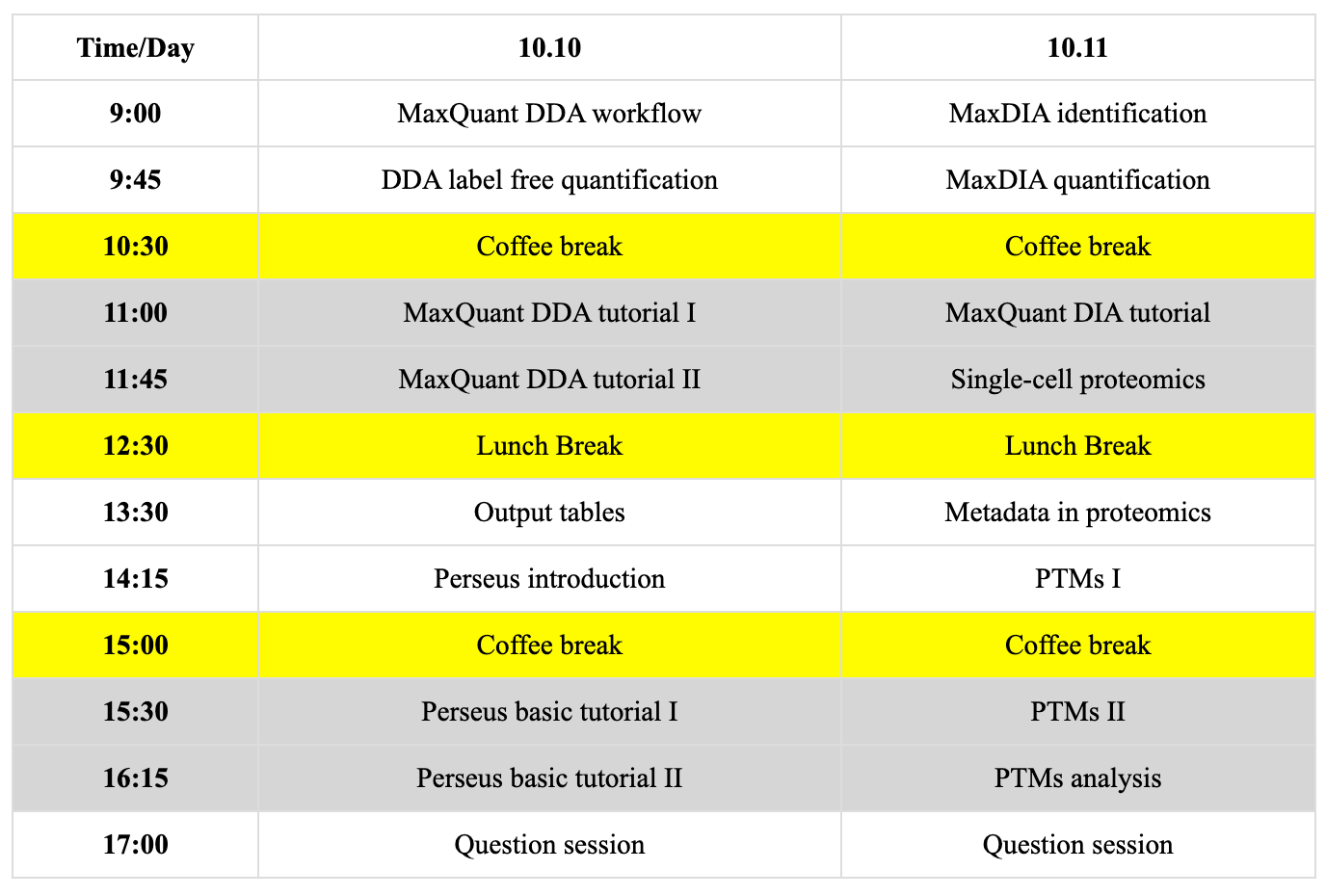
深度可视化蛋白质组学(Deep Visual Proteomics, DVP)是一项Mann实验室(哥本哈根以及慕尼黑实验室)开发的前沿的空间蛋白质组学技术。它融合了高分辨率显微成像、人工智能驱动的细胞分割、激光显微切割(LMD)以及高灵敏质谱分析。该方法能够在单细胞甚至亚细胞水平(可检测超过5000种蛋白质)实现深度蛋白质组解析,适用于福尔马林固定石蜡包埋(FFPE)及冷冻组织切片,并与病理形态学紧密结合。
自2022年首次发表以来,DVP已成功应用于多种疾病研究,在揭示空间分辨分子机制方面的独特优势。相关应用包括中毒性表皮坏死松解症(一种致命性皮肤疾病)、α1-抗胰蛋白酶缺乏症相关肝病,以及各种癌等研究。这些成果凸显了DVP在刻画组织异质性、免疫微环境及疾病进展方面的多样性与潜力为精准医学提供了重要的生物学见解。
本次培训课程由DVP技术的开发团队全面介绍DVP的技术流程——从样本制备到数据分析——并结合实际案例展示其在肿瘤学与转化医学中的应用。培训时间为09:00至13:30,期间安排工作午餐与现场答疑。
Deep Visual Proteomics (DVP) is a cutting-edge spatial proteomics technology developed by Mann’s groups in Copenhagen and Munich. It integrates high-resolution microscopy, AI-powered cell segmentation, laser microdissection (LMD), and high-sensitivity mass spectrometry. This approach enables in-depth proteomic profiling at single-cell and subcellular resolution (>5000 proteins), applicable to both FFPE and fresh frozen tissue sections, and closely linked with morphological pathology.
Since its first publication in 2022, DVP has been successfully applied to a broad spectrum of diseases, demonstrating its unique ability to reveal cell-type resolved molecular mechanisms, for example, toxic epidermal necrolysis, a lethal skin disease, alpha-1 antitrypsin deficiency liver disease, as well as studies in types of cancers. These cases showcase DVP’s versatility in characterizing tissue heterogeneity, immune microenvironment, and disease progression, providing valuable insights for precision medicine.
This workshop will cover the complete DVP workflow – from sample preparation to data analysis – and present real-world case studies demonstrating its applications in oncology and translational medicine. The program runs from 09:00 to 13:30, with a working lunch and Q&A.


♦ 2025 PEAKS 培训课程|AOHUPO&AOAPO&CNHUPO&π-HuB 2025大会系列活动
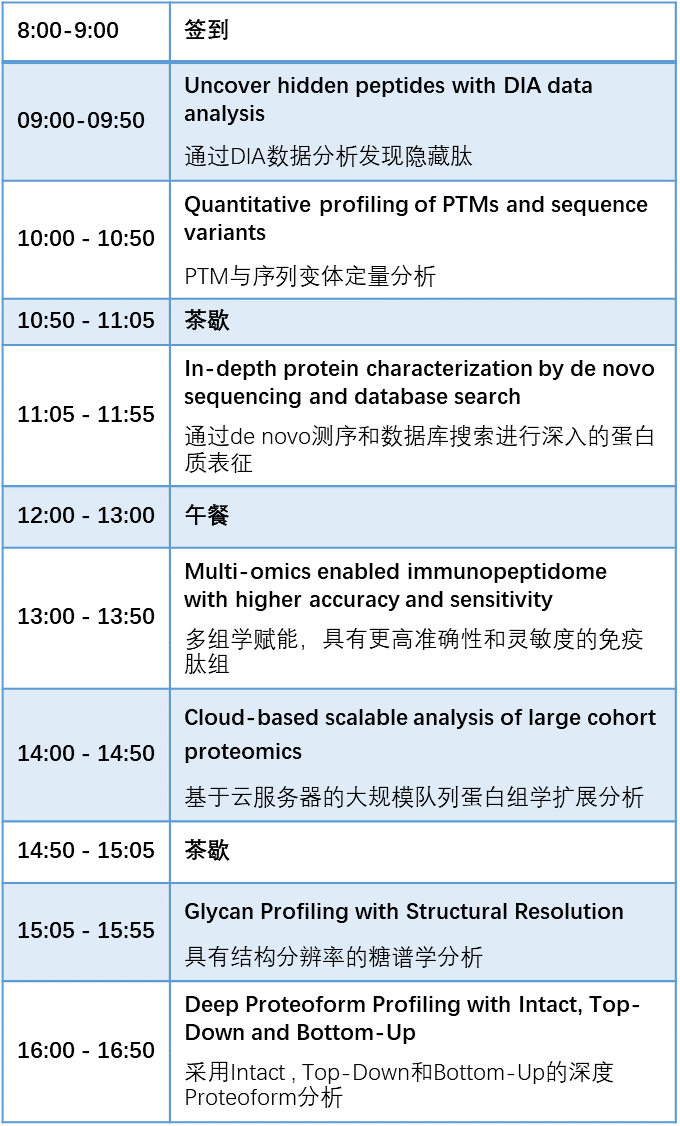
在2025年广州 CNHUPO/AOHUPO 联合大会上,我们举办本次为期两天的 PEAKS Training Workshop,将围绕 “探索蛋白质宇宙:迈向新生物学与精准医学” 的大会主题,深入探讨肽组与蛋白组”暗物质”领域。
在过去的几年中,大规模的蛋白质组学研究使我们能够开始在蛋白质水平上了解表型。与人类基因组中大约 20,000 个蛋白质编码基因相比,人体内有超过 100,000 种不同的蛋白质,并且由于可变剪接以及转录后和翻译后加工,还存在数十万种 proteoform。多项课题研究表明蛋白质形态比其相应的蛋白质能更好地描述蛋白质水平的生物学并且是更具体的分化指标。且人工智能(AI)模型的赋能带来了高通量蛋白质组学的革命。
本次为期两天的 PEAKS 培训课程,将介绍 AI 模型在高通量蛋白质组学中的应用;展示如何针对多种互补的质谱方法,结合数据库搜索和从头测序的数据分析,达到在proteoform水平上的深度覆盖。无论是对于刚刚进入AI赋能的蛋白质组领域的新手,还是有经验的大队列样本的蛋白质组学研究,都非常适合参加这个培训。
课程主要内容聚焦在:
1. 利用新型人工智能模型,从DIA数据中揭示暗肽组/蛋白质组,直接从复杂的DIA谱图中鉴定出肽序列变体和新的多肽,同时严格控制错误发现率。
2. 大规模队列项目数据分析的自动化,以获得深入的蛋白质组学和蛋白质基因组学见解,从而提高发现通量并开发基于质谱的靶向技术。
3. 演示如何整合完整蛋白质分析、自上而下和自下而上的方法,以实现全面的蛋白质组表征,包括可变剪接、翻译后修饰和变体。
For the 2025 CNHUPO/AOHUPO Joint Conference in Guangzhou, the PEAKS two-day workshop will revolve around the theme Navigating the Protein Universe: Toward New Biology and Precision Medicine, exploring the dark proteome and peptidome in depth.
Over the past few years, large-scale proteomics efforts have allowed us to begin to understand phenotype at a protein level. In contrast to about 20,000 protein-coding genes in the human genome, there are over 100,000 different proteins and, as a result of alternative splicing, hundreds of thousands of protein isoforms (variants) in the human body. Proteoforms better describe protein-level biology and are more specific indicators of differentiation than their corresponding proteins. It comes the revolution in high-throughput proteomics with artificial intelligence (AI) models.
In PEAKS Online training workshop, we are going to introduce AI models for high-throughput proteomics with deep coverage at proteoform level, by integrate alternative MS approaches with complementary database search and de novo sequencing data analysis. This training is highly suitable for both newcomers who have just entered the AI-empowered proteomics field and experienced researchers engaged in proteomics studies of large cohort samples.
Highlights of the training course:
1. Uncover dark peptidome/proteome from DIA data by a novel AI model to identify peptide sequence variants and novel peptides directly from complex DIA spectra while rigorously controlling the false discovery rate.
2. Learn the automation of data analysis of large-scale cohort project to obtain in-depth proteomics and proteogenomics insights for throughput of discovery and targeted mass-spectrometry-based technologies
3. Demo how to integrate intact protein analysis, Top-Down and Bottom-Up approaches, to enable comprehensive proteoform characterization, including alternative splicing, modifications, and variants.
♦ 2025 AI for Protein Science培训课程|AOHUPO&AOAPO&CNHUPO&π-HuB 大会
AI for Protein Science课程旨在系统介绍人工智能如何变革蛋白质科学研究的各个方面,专注于人工智能技术在蛋白质科学领域的深度应用。讲座内容包括蛋白质组数据资源及多组学分析策略介绍、临床队列蛋白质谱定量统计与注释、SpectraAI、大模型在蛋白质翻译后修饰研究中的应用、组学大模型前沿技术综述。培训时间为9:00至12:30。
AI for Protein Science aims to systematically introduce how artificial intelligence is transforming various aspects of protein science research, with a focus on the in-depth application of AI technologies in the field of protein science.
Lecture topics include: Introduction to proteomic data resources and multi-omics analysis strategies, statistical analysis and annotation of clinical cohort proteomics quantification data, SpectraAI, the application of large models in protein post-translational modification research, and a review of frontier technologies in omics-based large models. The program runs from 9:00 to 12:30.

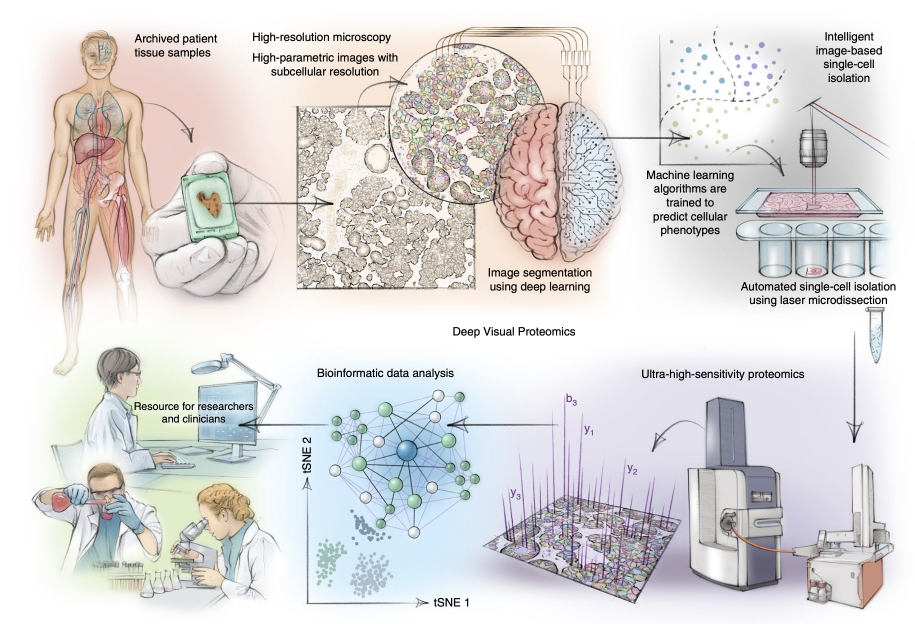
♦ 2025 MaxQuant 软件培训课程|AOHUPO&AOAPO&CNHUPO&π-HuB 2025大会系列活动
Juergen Cox团队的MaxQuant培训课程是基于质谱的蛋白质组学数据进行数据分析的理论与实践教学,于 2009 年首次举办,每年都有数百位参与者。
在广州的两天培训课程由介绍理论的讲座和结合真实数据集的软件实操组成。软件的开发者们会展示如何用最新版本的MaxQuant进行DDA和DIA数据分析,以及如何用Perseus继续进行下游的数据分析,如何进行PTM数据分析和单细胞蛋白质组等数据分析。每天课后设置了问答环节来解决培训时没有回答的问题。无论是刚刚进入蛋白质组领域的新手还是有经验的研究者,都非常适合参加这个培训。
The MaxQuant workshop in Dr. Juergen Cox’s group provides hands-on training in the computational analysis of proteomics data generated by modern mass spectrometers. Since the first MaxQuant Summer School in 2009, this annual event has attracted hundreds of participants every year.
The two-day workshop in Guangzhou will consist of lectures that focus on theory and tutorials that focus on how to use the software with the dataset we provide. The developers of the software will show the DDA and DIA analysis withlatest MaxQuant and the downstream analysis with Perseus. We’ll also introduce some advanced analysis such as PTMs, single cell proteomics, etc. There will also be a Q&A session at the end of the day to answer any questions not covered at the end of the presentation. The workshop is suitable for both beginners and experienced proteomics researchers.